FGF pathway map
|
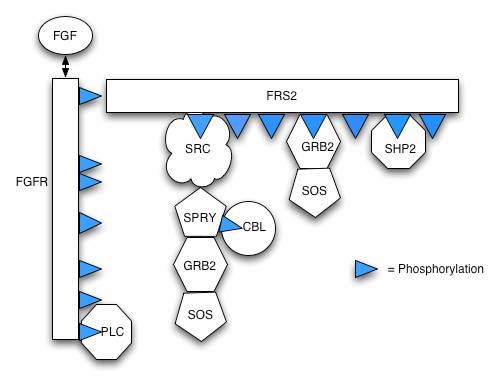
| Pi-calculus model containing processes FGF, FGFR, FRS2, Plc, Src, Spry, Cbl, Grb, Shp and Sos (model extension with Src positive feedback here)
| |
|
|
Reactions
| |
These reactions approximate the behaviour of the FGF pathway (see SBML for the full model).
|
1. FGF binds to FGFR
FGF+FGFR ↔ FGFR:FGF (kon=5e+6 M-1s-1, koff=5e-3 s-1)
2. Whilst FGFR:FGF exists
FGFR Y653/Y654 → FGFR Y653P/Y654P (kcat=0.013s-1)
3. When FGFR Y653P/Y654P
FGFR Y766 → FGFR Y766P (kcat=0.004s-1)
4. FGFR binds FRS2
FGFR+ FRS2 ↔ FGFR:FRS2 (kon=2.5e+6 M-1s-1, koff=5e-2 s-1)
5. When FGFR Y653P/Y654P and FGFR:FRS2
FRS2 Y196 → FRS2 Y196P (kcat=0.005s-1)
FRS2 Y306 → FRS2 Y306P (kcat=0.005s-1)
FRS2 Y471 → FRS2 Y471P (kcat=0.005s-1)
6. Reverse when Shp2 bound to FRS2:
FRS2 Y196P → FRS2 Y196 (kcat=12s-1)
FRS2 Y306P → FRS2 Y306 (kcat=12s-1)
FRS2 Y471P → FRS2 Y471 (kcat=12s-1)
7. FRS2 effectors bind phosphoFRS2:
Src+FRS2 Y196P ↔ Src:FRS2 Y196P (kon=2.5e+6 M-1s-1, koff=5e-2 s-1)
Grb2+FRS2 Y306P ↔ Grb2:FRS2 Y306P (kon=2.5e+6 M-1s-1, koff=5e-2 s-1)
Shp2+FRS2 Y471P ↔ Shp2:FRS2 Y471P (kon=2.5e+6 M-1s-1, koff=5e-2 s-1)
8. When Src:FRS2 we relocate/remove
Src:FRS2 → relocate out (t1/2 = 15min)
9. When Plc:FGFR it degrades FGFR
PLC+FGFR Y766 ↔ PLC:FGFR Y766 (kon=2.5e+6 M-1s-1, koff=5e-2 s-1)
PLC:FGFR Y766 → degFGFR (t1/2 = 60 min)
10. Spry appears in time dependent manner:
→ Spry (t = 20min)
11. Spry binds Src and is phosphorylated:
Spry+Src ↔ Spry Y55:Src (kon=1e+5 M-1s-1, koff=1e-2 s-1)
Spry Y55:Src → Spry Y55P:Src (kcat=10s-1)
Spry Y55P+Src ↔ Spry Y55P:Src (kon=1e+5 M-1s-1, koff=1e-4 s-1)
Spry Y55P+Cbl ↔ Spry Y55P:Cbl (kon=1e+5 M-1s-1, koff=1e-4 s-1)
Spry Y55P+Grb2 ↔ Spry Y55P:Grb2 (kon=2.5e+6 M-1s-1, koff=5e-2 s-1)
12. phosphoSpry binds Cbl which degrades/removes FRS2
Spry Y55P:Cbl+FRS2-Ubi → FRS2-Ubi (t1/2 = 25min)
13. Spry is dephosphorylated by Shp2: (when Shp2 bound to FRS2)
Spry Y55P → Spry Y55 (kcat=12s-1)
14. Grb2 binds Sos
Grb2+Sos ↔ Grb2:Sos (kon=1e+5 M-1s-1, koff=1e-4 s-1)
|
|
|
